Abstract
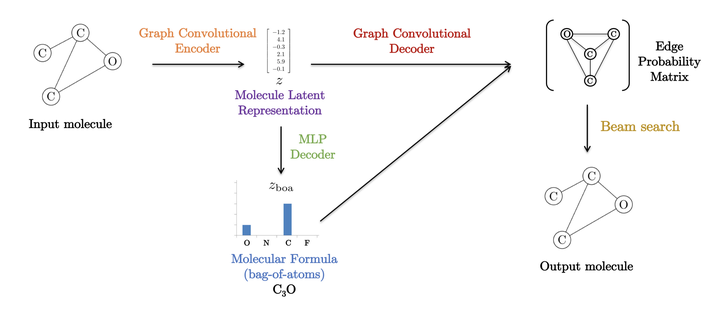
We propose a simple auto-encoder framework for molecule generation. The molecular graph is first encoded into a continuous latent representation , which is then decoded back to a molecule. The encoding process is easy, but the decoding process remains challenging. In this work, we introduce a simple two-step decoding process. In a first step, a fully connected neural network uses the latent vector to produce a molecular formula, for example CO (one carbon and two oxygen atoms). In a second step, a graph convolutional neural network uses the same latent vector to place bounds between the atoms that were produced in the first step (for example a double bound will be placed between the carbon and each of the oxygens). This two-step process, in which a bag of atoms is first generated, and then assembled, provides a simple framework that allows us to develop an efficient molecule auto-encoder. Numerical experiments on basic tasks such as novelty, uniqueness, validity and optimized chemical property for the 250k ZINC molecules demonstrate the performances of the proposed system. Particularly, we achieve the highest reconstruction rate of 90.5%, improving the previous rate of 76.7%. We also report the best property improvement results when optimization is constrained by the molecular distance between the original and generated molecules.